Someone asked me why some Morpholinos usually cause exon skips and others cause intron inclusions...
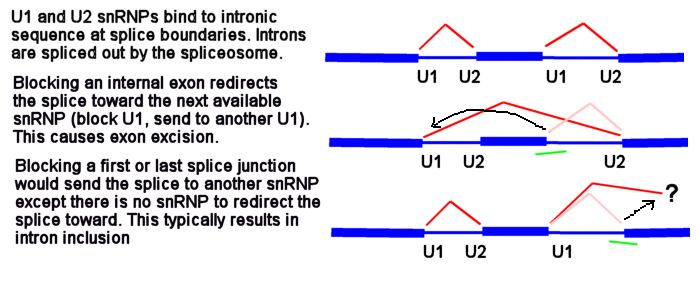
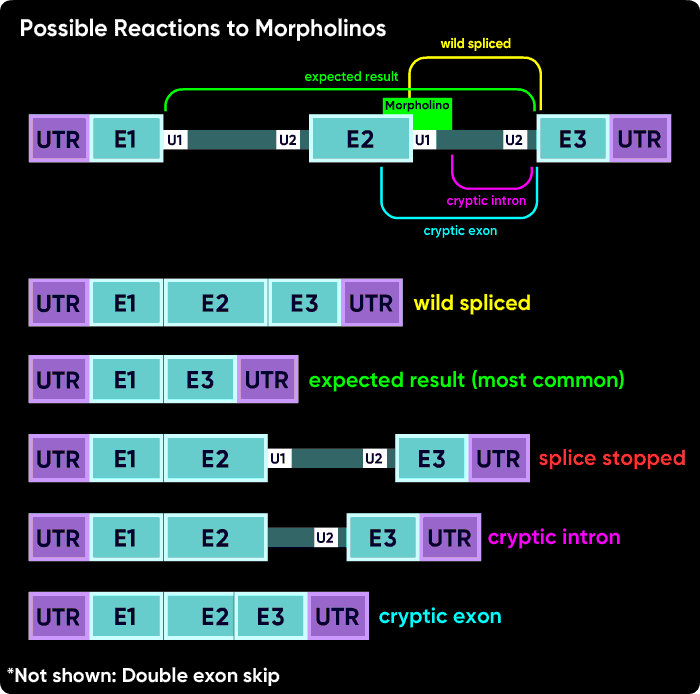
U1 and U2 (or U11 and U12) snRNPs mark the positions on pre-mRNA of the splice junctions for the spliceosome. There is a U1 snRNP that binds in the intron near the e2i2 junction and a U2 snRNP that binds on the other side of the intron at the i2e3 junction.
Figure 1
Morpholinos targeting splicing are usually targeted to block the binding sites of these snRNPs. Consider the splice junctions of the second intron. If you block the U2 snRNP binding site with an i1e2 oligo, the spliceosome will match the U1 snRNP at i1e2 with the next available U2 snRNP downstream, which is bound at the i3e3 junction. This splices the e1i1 junction to the i2e3 junction, eliminating i1, e2 and i2.
Figure 2

Now consider what happens if you block the e1i1 junction with a Morpholino. The U1 can't bind to its e1i1 site, but the U2 at the i1e2 site has no other upstream U1 it can be redirected toward to make a splice. In this case, the usual result is for no splicing to occur at i1; that is, you see an i1 inclusion in the mature mRNA.
Figure 3
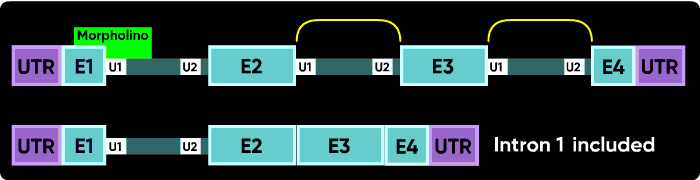
Because the very first (e1i1) and very last splice sites have no splice sites past them to which splicing can be redirected, blocking them causes a failure to splice (intron inclusion). Blocking any of the internal splice boundaries (i1e2, e2i2, i2e3, etc.) usually causes the adjacent exon to be eliminated from the mature mRNA (or, in the common jargon, causes the exon to be skipped). Here is an illustration of intron inclusion resulting from blocking the last splice junction.
Figure 4
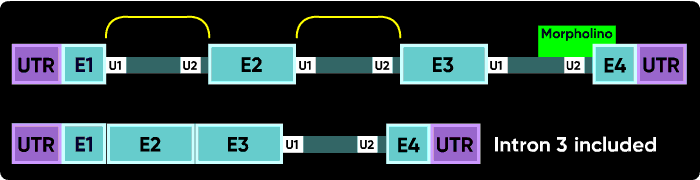
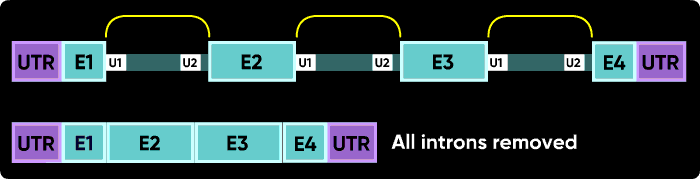
Other things can happen. Activation of cryptic splice sites can cause partial exon excision or partial intron inclusion. Sometimes an oligo causes a double exon skip. Another possible outcome is a failure to splice of an internal intron, causing its inclusion in the mature mRNA. However, the most common outcomes are a intron inclusion for the first and last splice sites and an exon excision (skip) for all other splice junctions.
Figure 5
An entirely different approach to modulating splicing is to block the binding sites of splice regulatory proteins (splice enhancers, splice suppressors). I'm mentioning it here, but not discussing it more.
Typically splice-modifying oligo efficacy is tested with reverse-transcript PCR followed by gel electrophoresis of the PCR product. It can take several primer pairs to determine what happened. Sometimes putting a primer in each of the exons adjacent to the target can reveal a clean excision of the targeted exon. To detect the exon 2 skip in the second figure above, you would typically put a primer in exon 1 and another in exon 3. Other common approaches are to move a primer an additional exon away to look for a double-exon-skip or to place a primer in intronic sequence to detect an intron inclusion. For cryptic splice site activation where the cryptic site is close to the wild-spliced site, sequencing the PCR product can reveal small changes. When choosing primers, be sure that you plan to make RT-PCR products of a hundred bases or so when the exon is missing; that way the RT-PCR product is long enough it can take up plenty of an intercalating fluorescent dye and be readily visible on the gel.
If you do not see an obvious single-exon-skip by looking at the RT-PCR product on an electrophoretic gel, one of these alternative splicing outcomes might be the reason (but the most likely outcome is the single-exon skip). Load the gel lightly and watch for dimming of the wild-spliced band in the MO-treated vs. control lane (normalized against a housekeeping gene band intensity). If the band is over-saturated with too much RNA , it can be harder to see the dimming of the band if some of the transcript is diverted to a different splicing fate, but if you have a lightly-loaded gel then it should be obvious if you lose some band intensity (normalized against a housekeeping gene). If nonsense-mediated decay is occurring, you might not see much or any mass-shifted band from splice-modification, but you should see the wild-spliced band become more dim.
One more time but drawn a bit differently - this is why blocking an internal splice site usually causes exon excision, but blocking the first or last splice site usually causes intron inclusion.
Figure 6